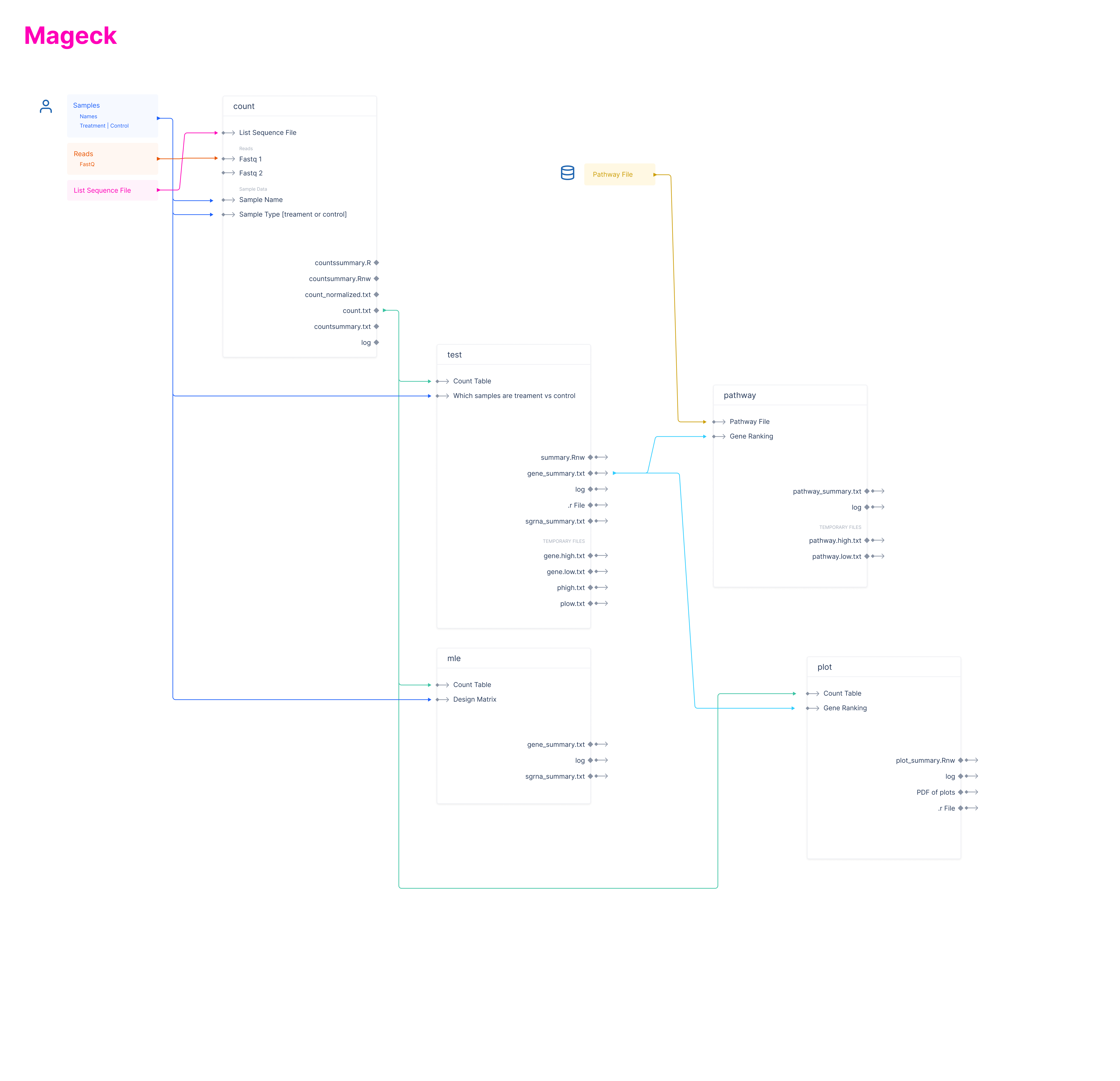
How to run MAGeCK on Latch
On Latch we have split up the MAGeCK workflow into its subcommands to be run. These are:MAGeCK - Count
MAGeCK - Test
MAGeCK - MLE
MAGeCK - Pathway
MAGeCK - Plot
mageck-subcommand-input-output-graph.pdf
1284.5 KB
Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout
