Opening Files in IGV
From Latch Data
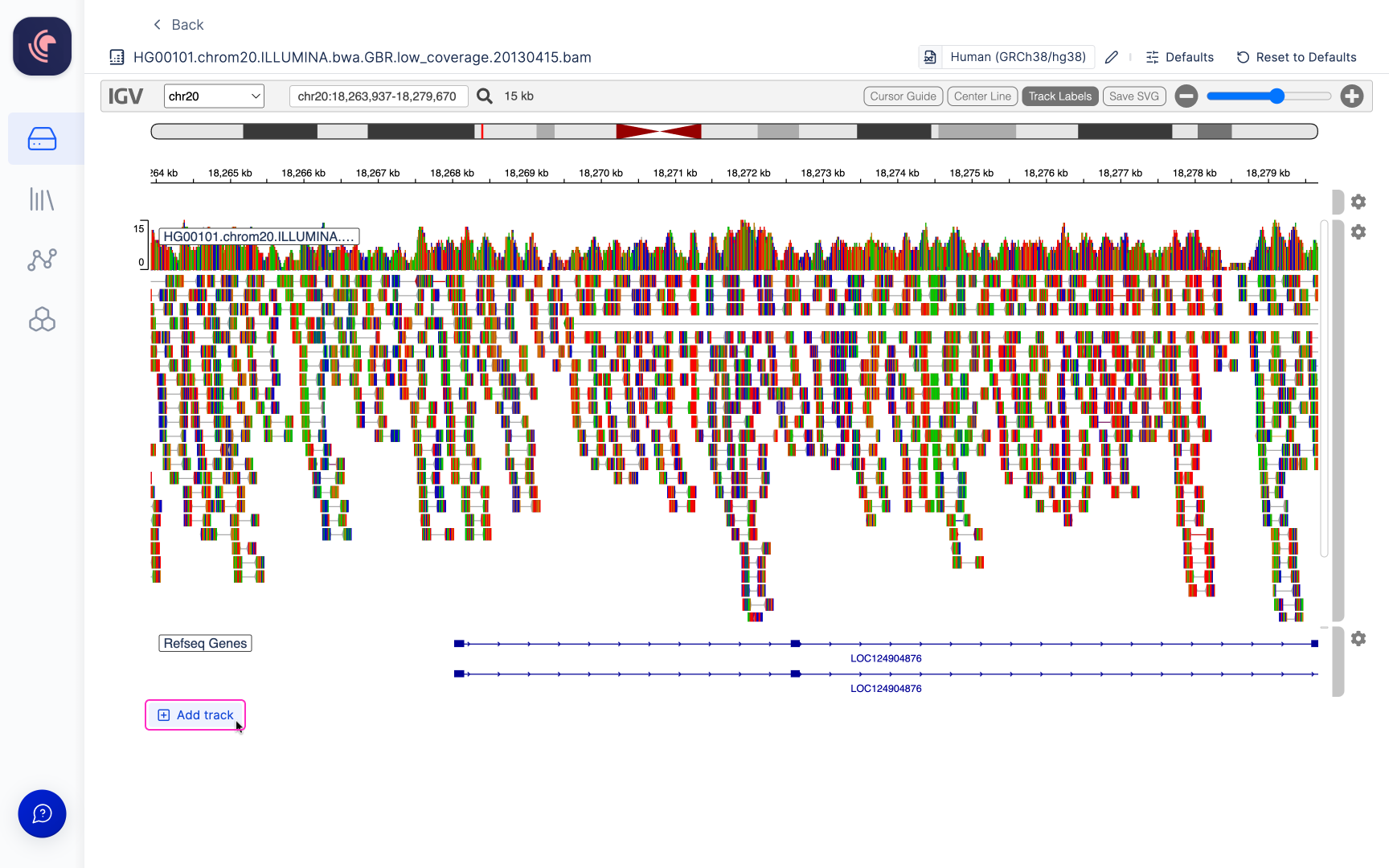
Nucleotide aequence (Fasta) and alignment files (BAM) files can be opened in the IGV browser from Latch Data. Simply double click a file to open it.
BAM Aligment Files
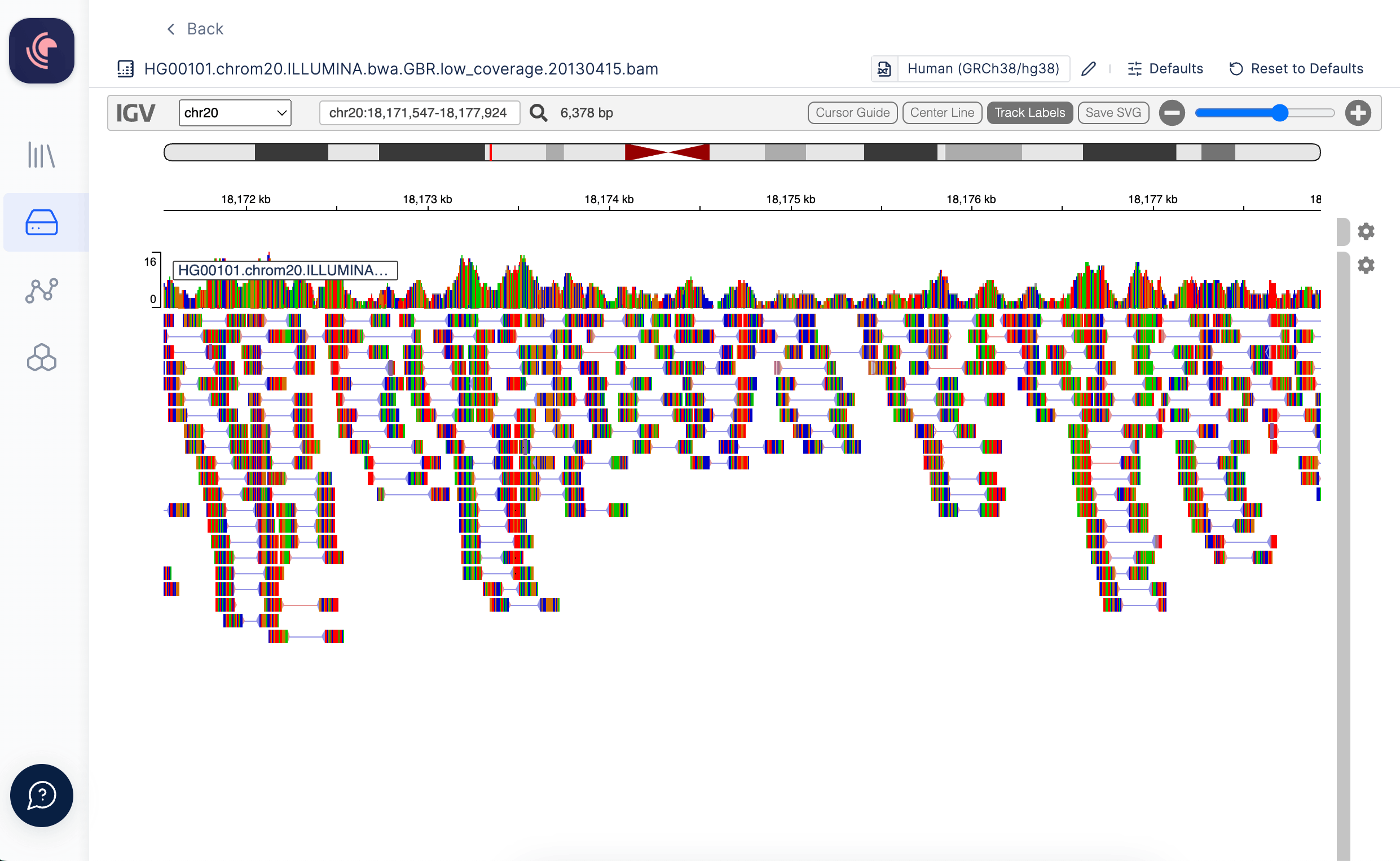
Alignment files (BAM) hosted on our platform are linked with an automatically generated index file (BAI), which is utilized for when opened in the Integrative Genomics Viewer (IGV). In order to view alignment files within IGV, the reference genome to which the data has been aligned must be specified. Users have the option to choose from a selection of commonly employed genomes available on IGV (a list of these options can be found here), or alternatively, you can select your own reference genomes stored within Latch Data. The IGV browser within Latch presently supports reference genome files in the .fasta, .fa, and .fna file formats.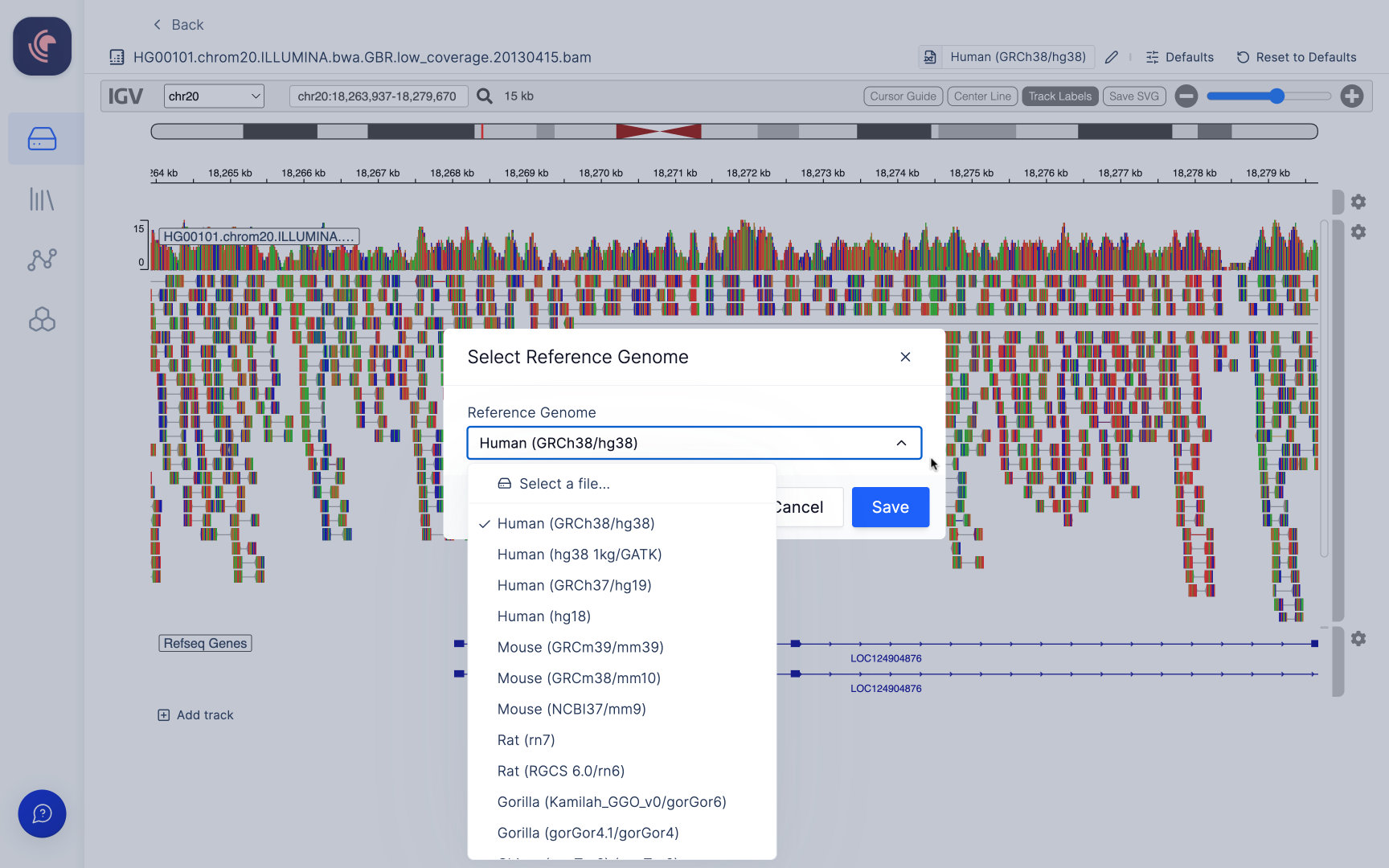
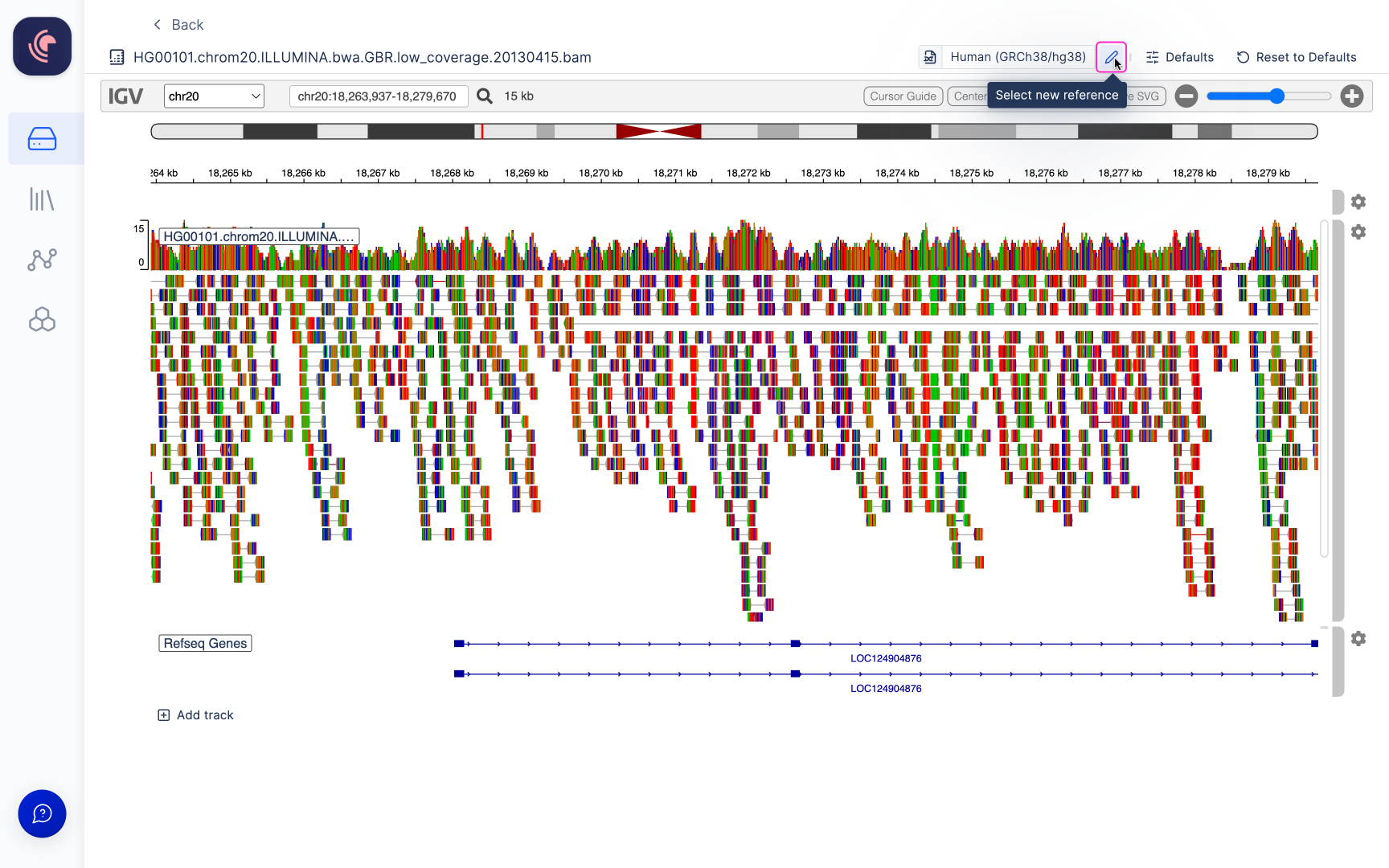
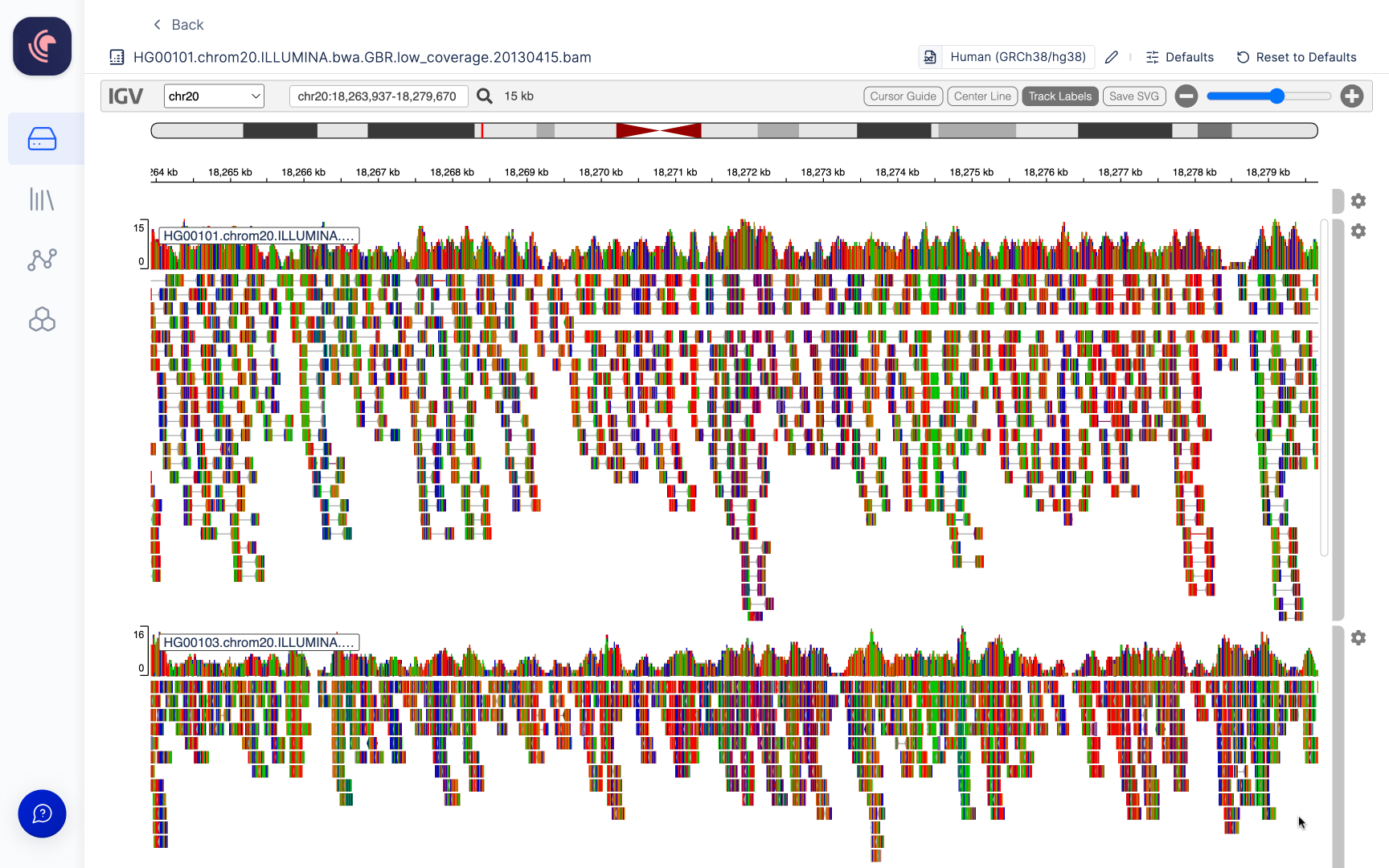
Adding additional tracks
Multiple BAM files can be opened in the viewer. To add a track scroll to the bottom of the last track displayed and click the+ Add Track button.