Quick Start
Inputs
Any .fastq or .fastq.gz file.Outputs
You’ll see an interactive .html report that looks like this.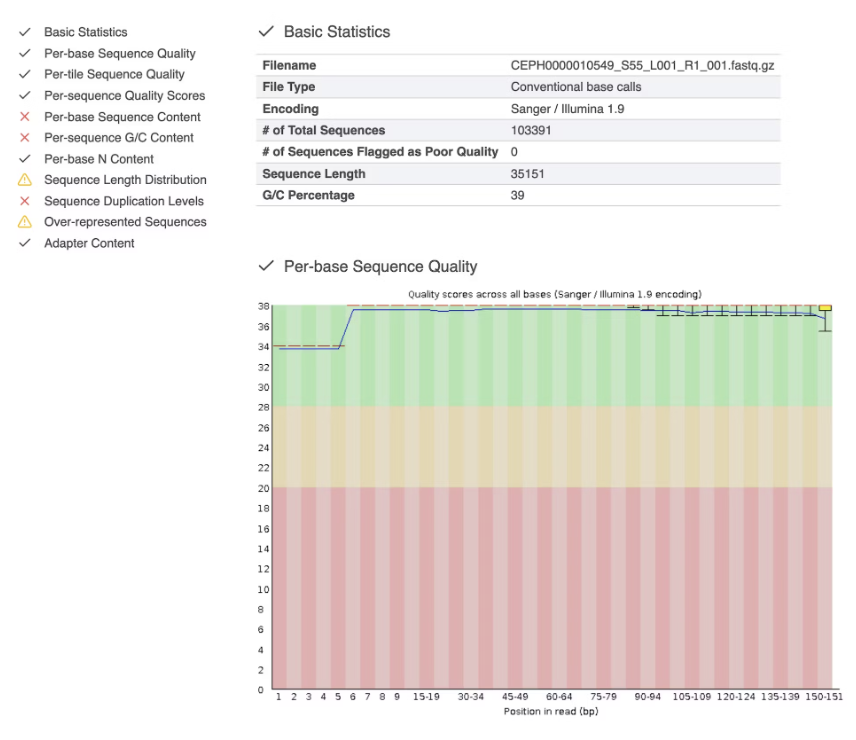
FastQC aims to provide a simple way to do some quality control checks on raw sequence data coming from high throughput sequencing pipelines. It provides a modular set of analyses which you can use to give a quick impression of whether your data has any problems of which you should be aware before doing any further analysis.
