1: Select the H5AD file you want to visualize
First, navigate to the H5AD file that you want to visualize. If this is your first time on Latch, you will find an example H5AD file under the folderwelcome/scbrowser/scanpy-pbmc3k.h5ad
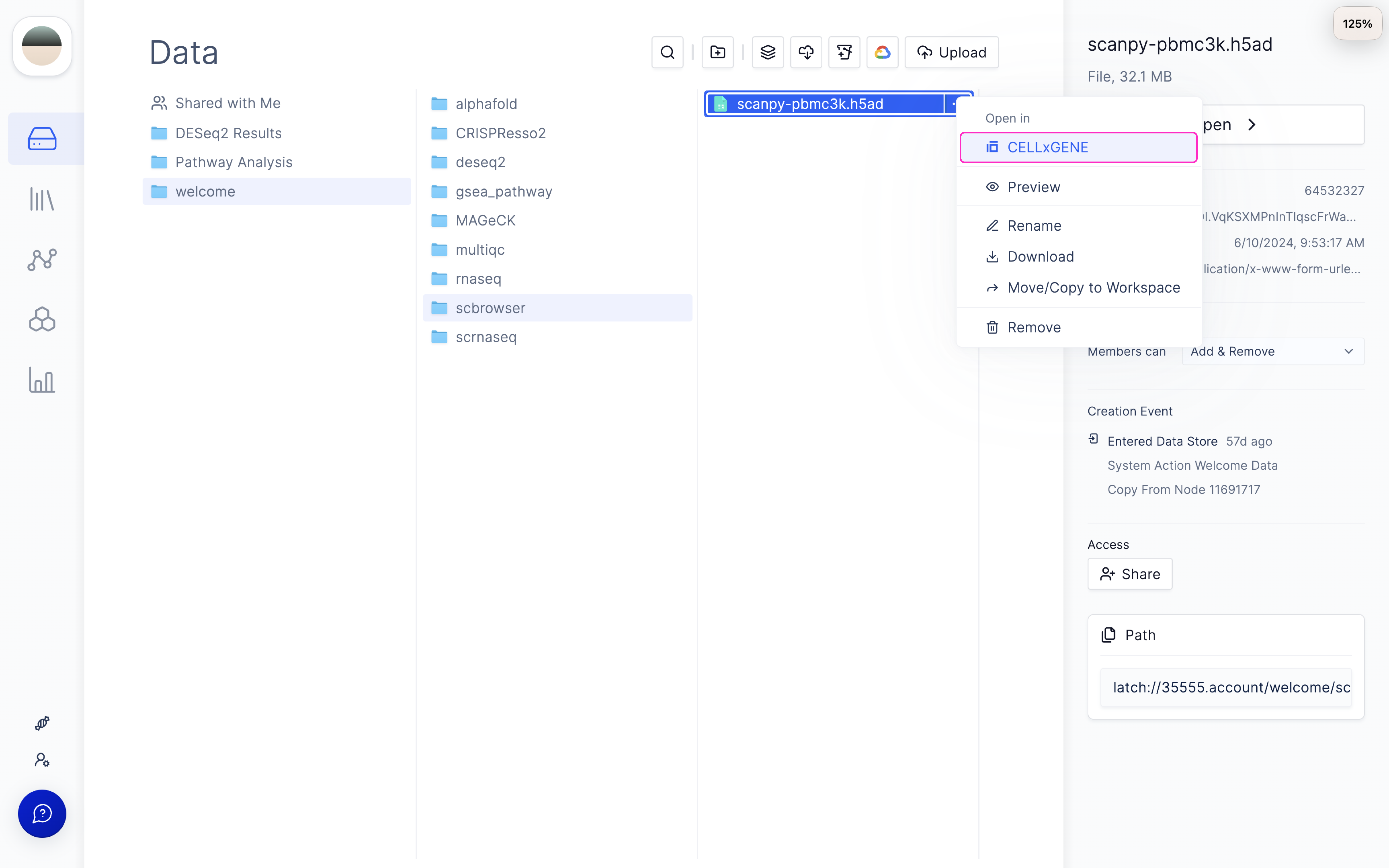
2: Start a new CELLxGENE Pod session
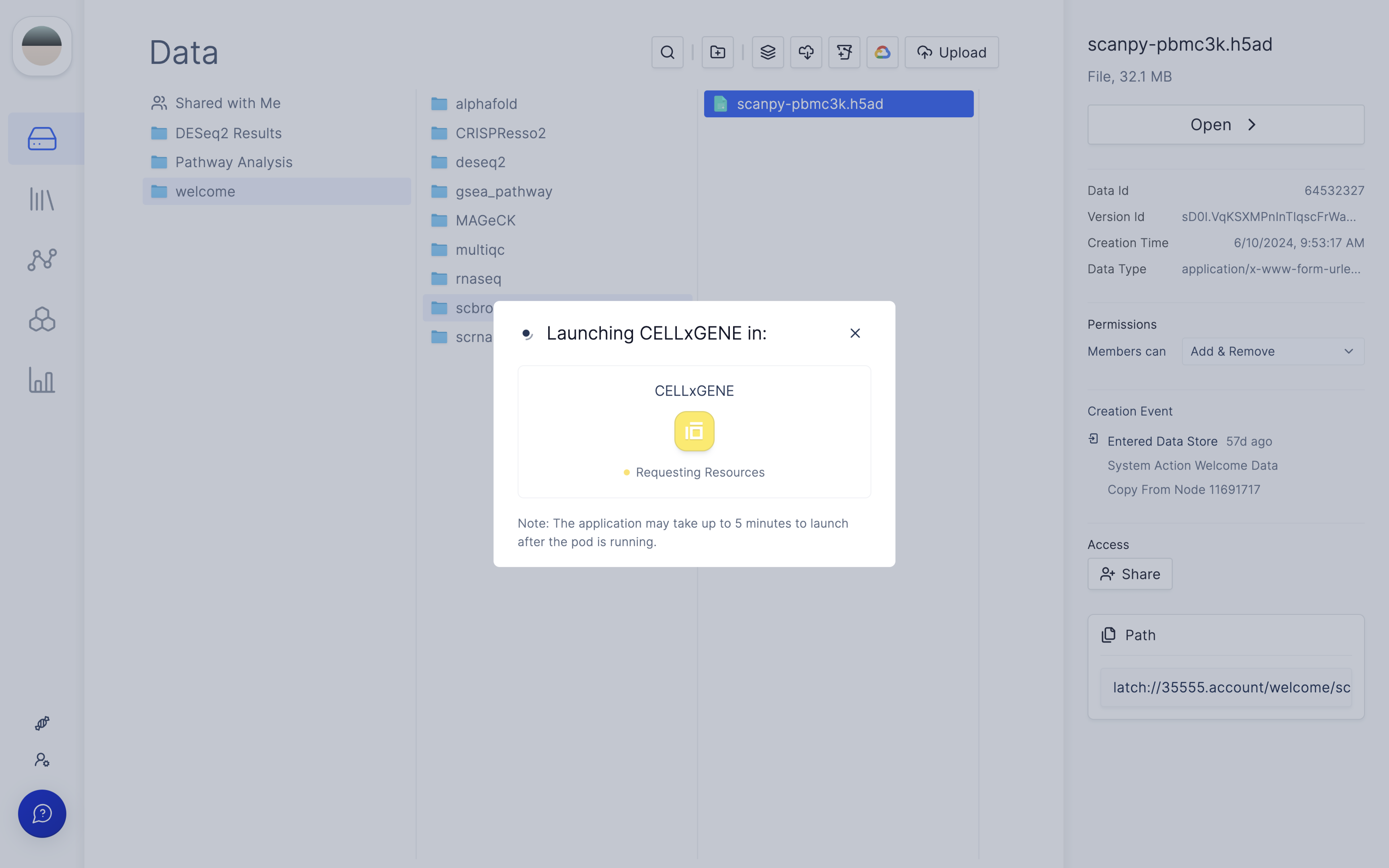
When you open CELLxGENE on the file for the first time, a new Latch Pod will be created.
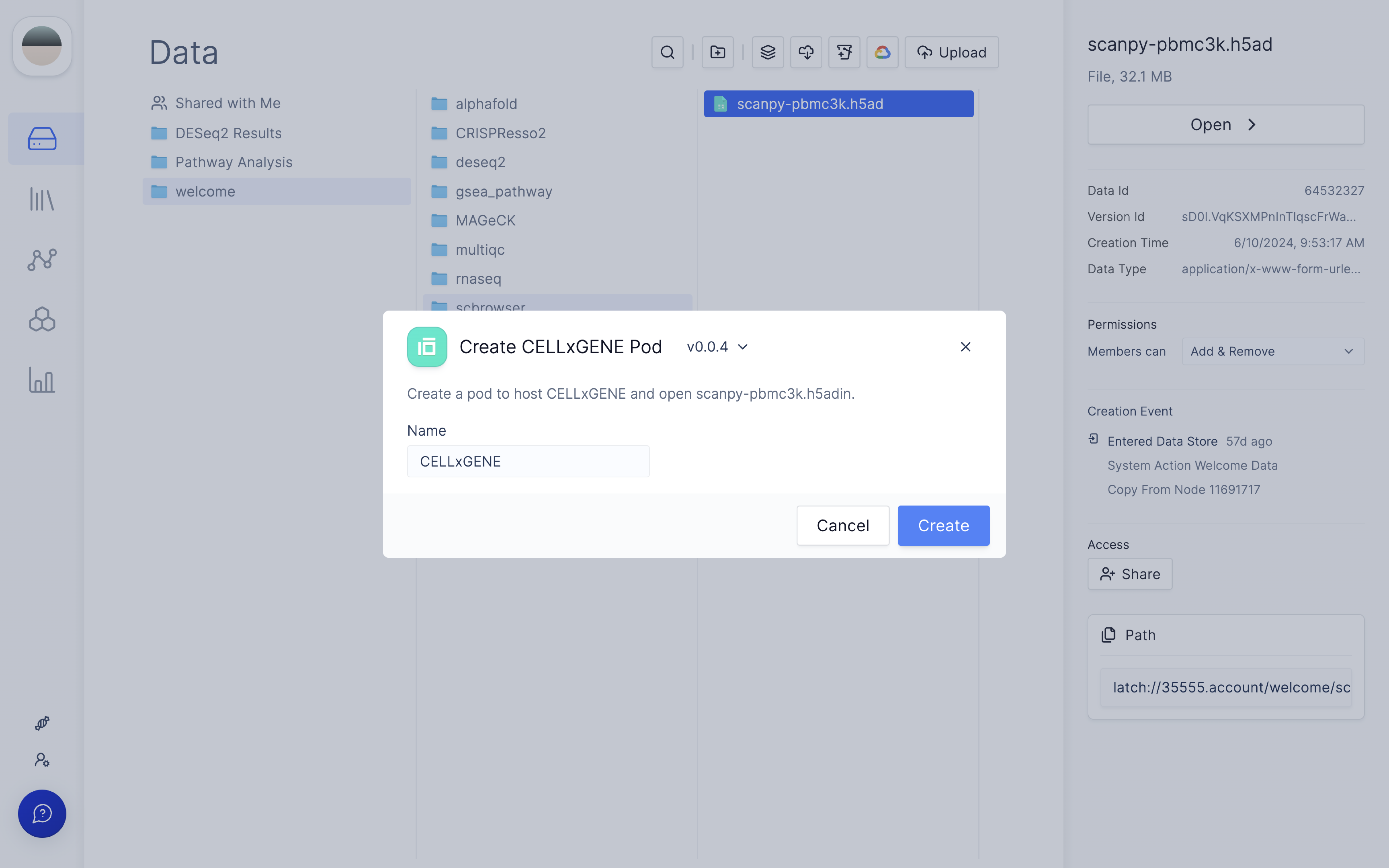
Once you click 'Create', you will see a status that indicates a Latch Pod is Launching.
FAQ: What contributes to the wait time?
FAQ: What contributes to the wait time?
- A computer instance is started with the default configuration of 8 CPUs and 32 GiB RAM.
- The necessary file is downloaded from Latch Data into the Pod.
- The
cellxgene launchcommand is automatically executed on the downloaded file.
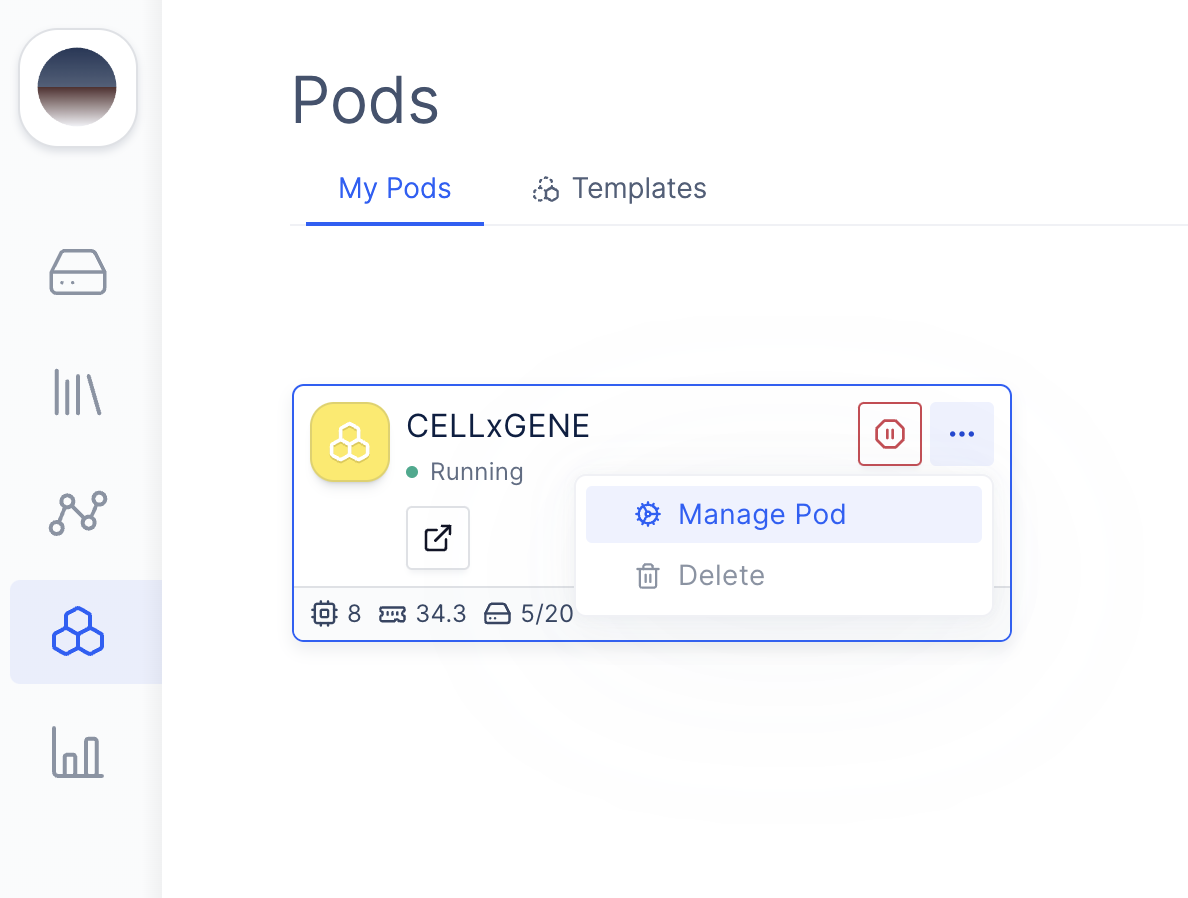
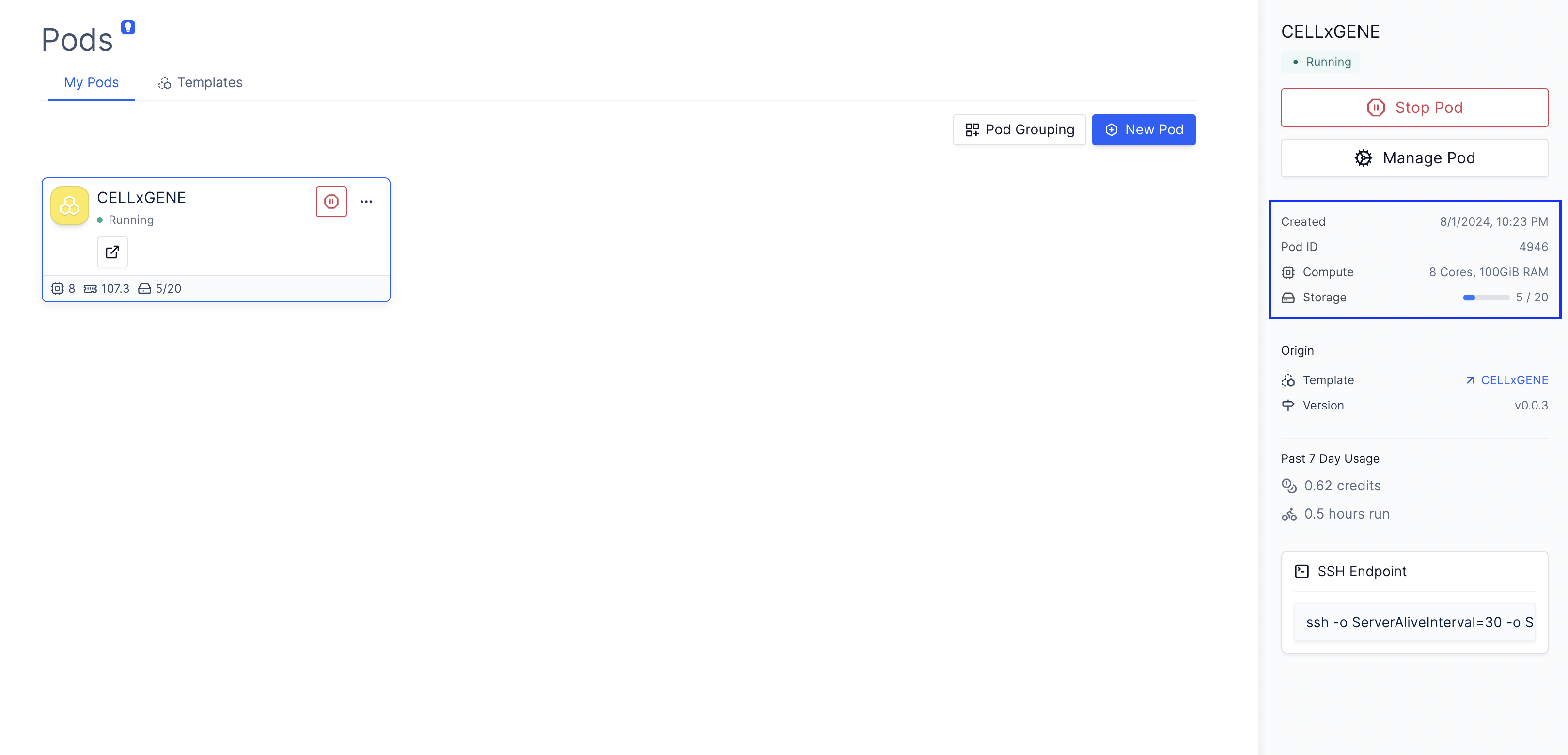
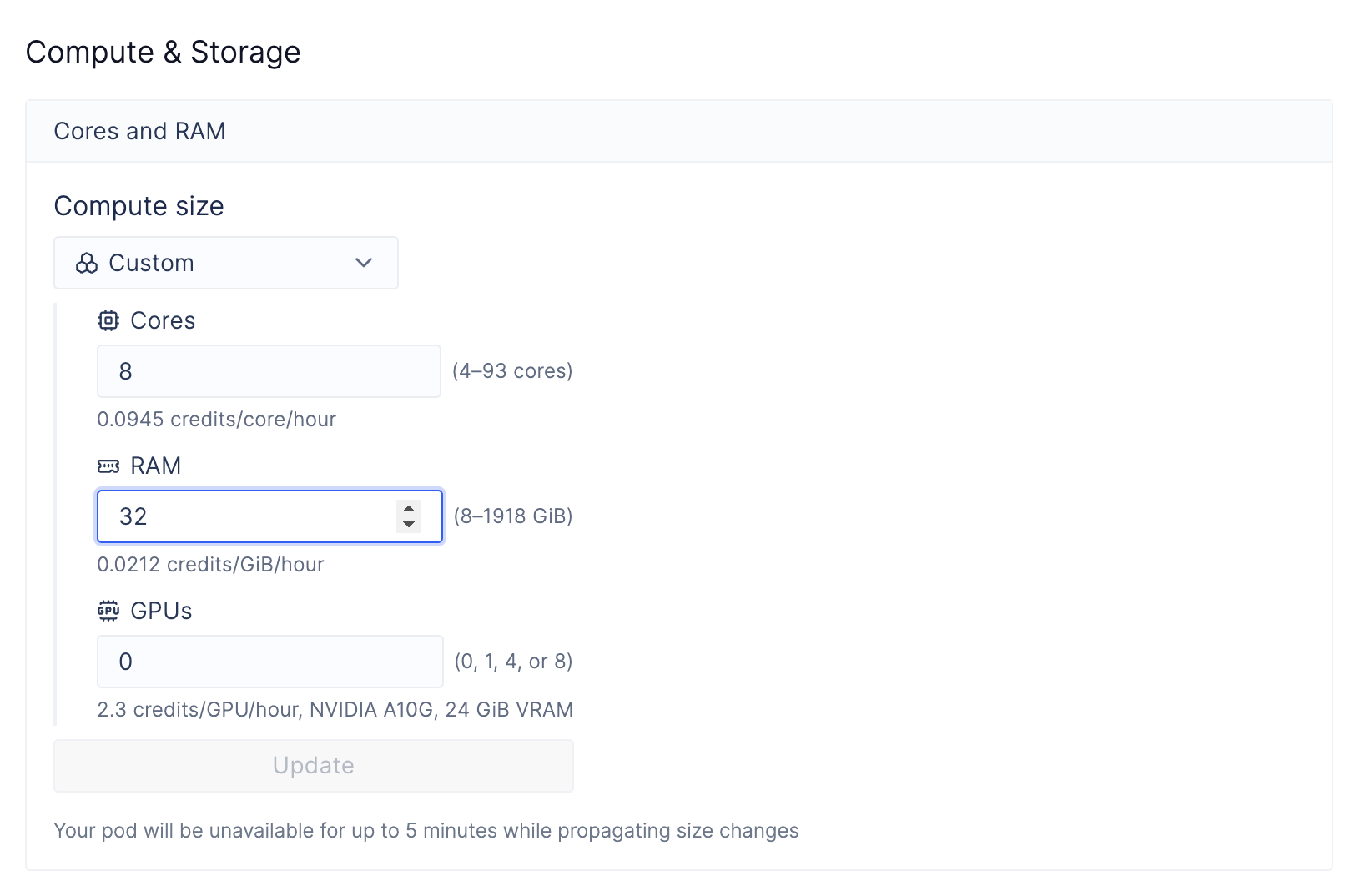
(Optional) 3: Change the compute resources for CELLxGENE
Sometimes, it’s often desirable for single-cell applications to increase the computer’s RAM for compute-intensive steps, such as computing TSNE and PCA, or running differential expression analyses. Because CELLxGENE is directly hosted on a Latch Pod that you have direct access to, changing the underlying resource profile is easy.Navigate to Latch Pods
Modify Compute
(Optional) 4: Turn off the Pod to save costs
Once you finish your analysis, you can go to the Pod and click the Stop button to shut down the Pod.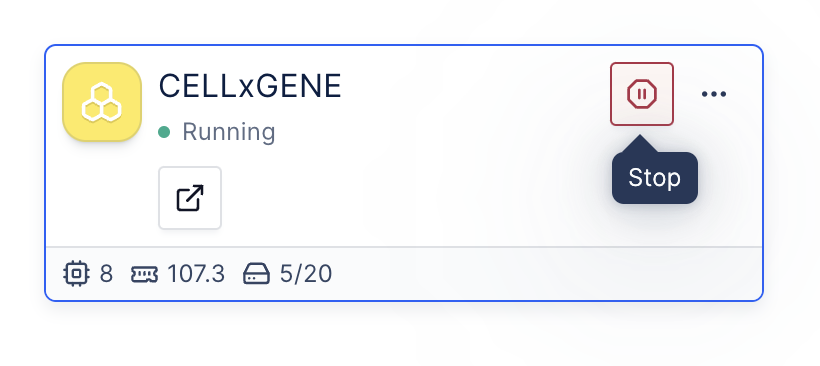
Frequently Asked Questions
Can I open multiple h5ad files per Pod?
Can I open multiple h5ad files per Pod?
(Advanced) I'm a developer and want to modify the startup script in my Pod when an h5ad file is selected. How do I do that?
(Advanced) I'm a developer and want to modify the startup script in my Pod when an h5ad file is selected. How do I do that?
/opt/latch/custom_app. You can use a code editor like vim or nano to modify the script.(Advanced) My Pod is taking unusually long to start (10+ minutes). How can I see the logs to understand what may be wrong?
(Advanced) My Pod is taking unusually long to start (10+ minutes). How can I see the logs to understand what may be wrong?