Overview
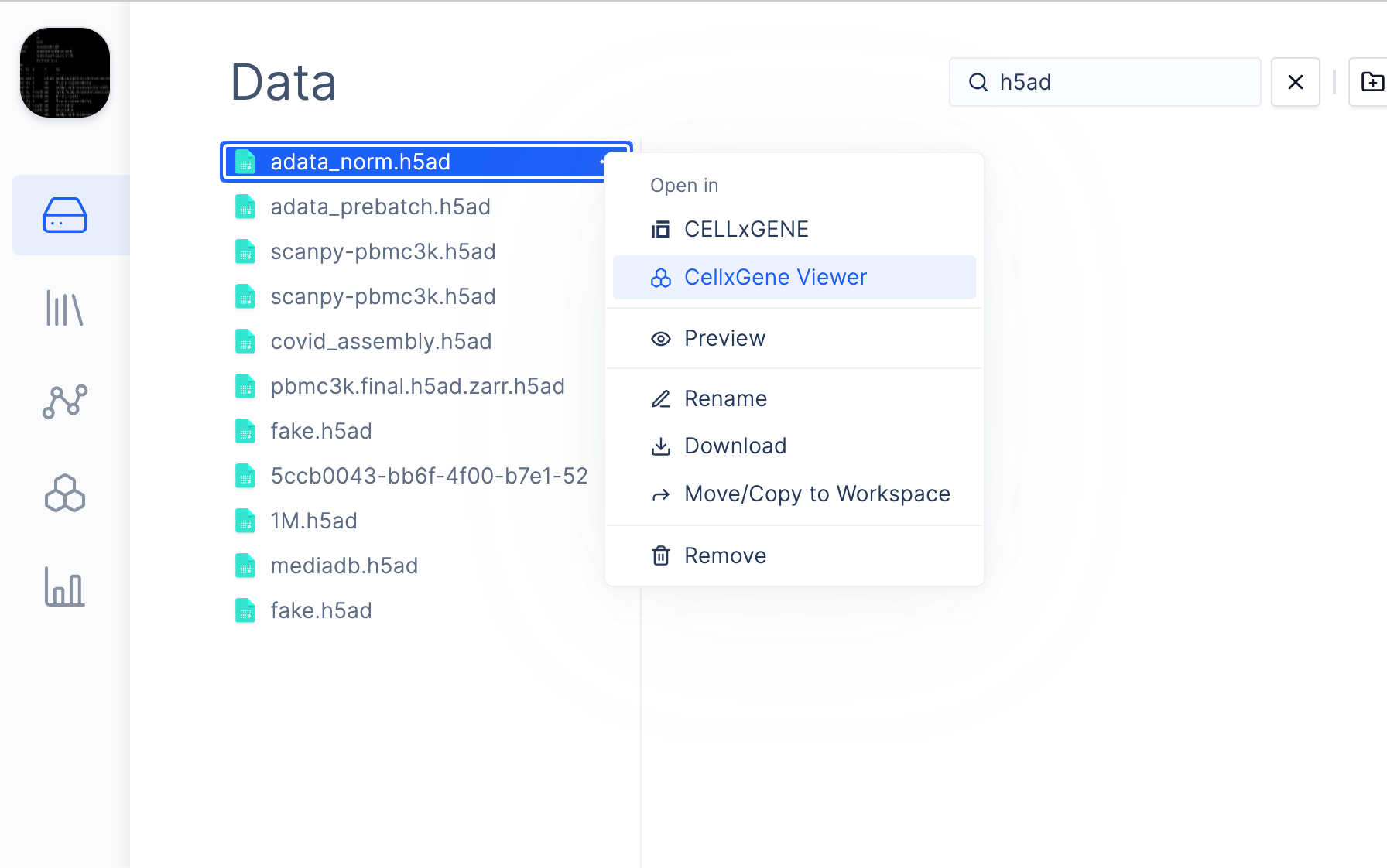
File viewers are Custom Apps built on top of Latch Pod Templates. These templates are configured to host a custom application that accepts a file path as a parameter and downloads that file for the application to access. In this example, we will be setting up cellxgene to view.h5ad files in Latch Data. Note that the
cellxgene file viewer is already available in Latch Data by default.
1. Set up your Custom Application
Follow the instructions here to setup a pod that hosts your custom application. For cellxgene, the launch script (/opt/latch/custom_app) looks like this:
pbmc3k.final.h5ad.zarr.h5ad. However, to
launch this application from the Latch Data UI, we need to pass in the file path of the file we
want to view.
To do so, add the following command to your bash script:
LDATA_NODE_ARG environment variable from the pod’s environment.
This variable is injected by the Latch backend and contains the ID of the
node that the user clicked on in Latch Data to launch the pod.
Now that we have the node ID, we can use latch cp to download the file to the pod and launch the application.
- First check if the
LDATA_NODE_ARGenvironment variable is set. If not, this pod was not launched from a Latch Data file so we start the application with a default file. - Since downloading large files can take a long time, we cache the file locally in the pod. If the file already exists, we use it instead of re-downloading.
- Delete the cache directory to avoid stale files from consuming disk space.
- Download the file to the pod using
latch cp. We download to a.tmpfile and then rename it to the target file to avoid partial downloads from corrupting the cache.
2. Create a Pod Template
You should now have a pod that takes in a Latch Data file as a parameter, downloads the file locally, and launches a custom application using that file. To make this pod available in Latch Data, you need to create a pod template from it. Follow the instructions here to create a pod template from the pod created in step 1.3. Configure the Custom File Viewer
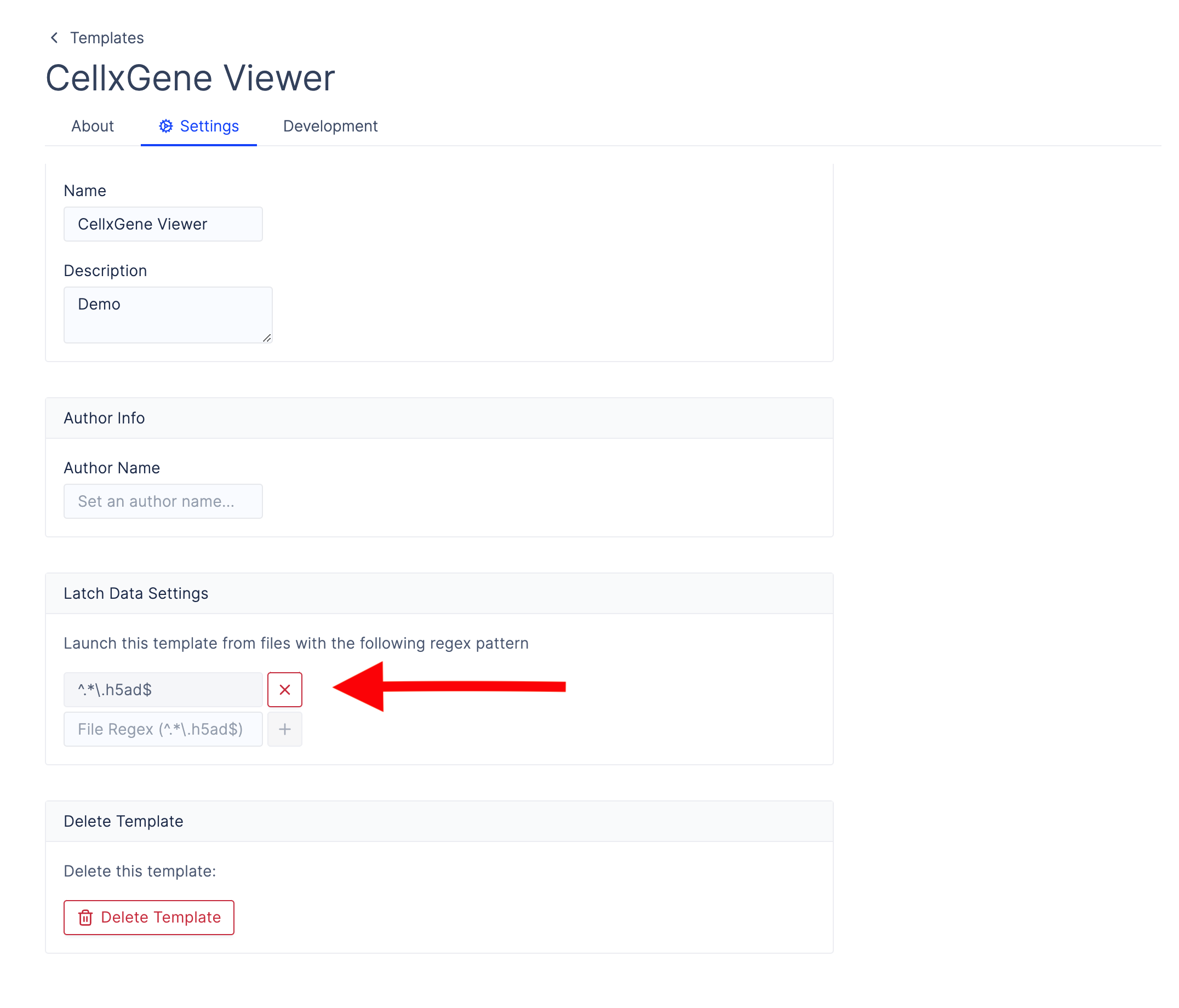
To configure Latch Data to open your pod template for a specific file pattern, go to the pod template settings page and add a regex pattern matching the files you want to open with your custom file viewer. For cellxgene, we want to restrict the viewer to.h5ad files. The regex pattern for this is ^.*\.h5ad$.
4. View Files
You should now be able to click on files in Latch Data containing the pattern defined in step 3 and launch a pod which will download the file and launch your application.