1. Create your first project
A few examples of projects:
A few examples of projects:
Discover optimal antibody candidates through multiple rounds of phage display assays.
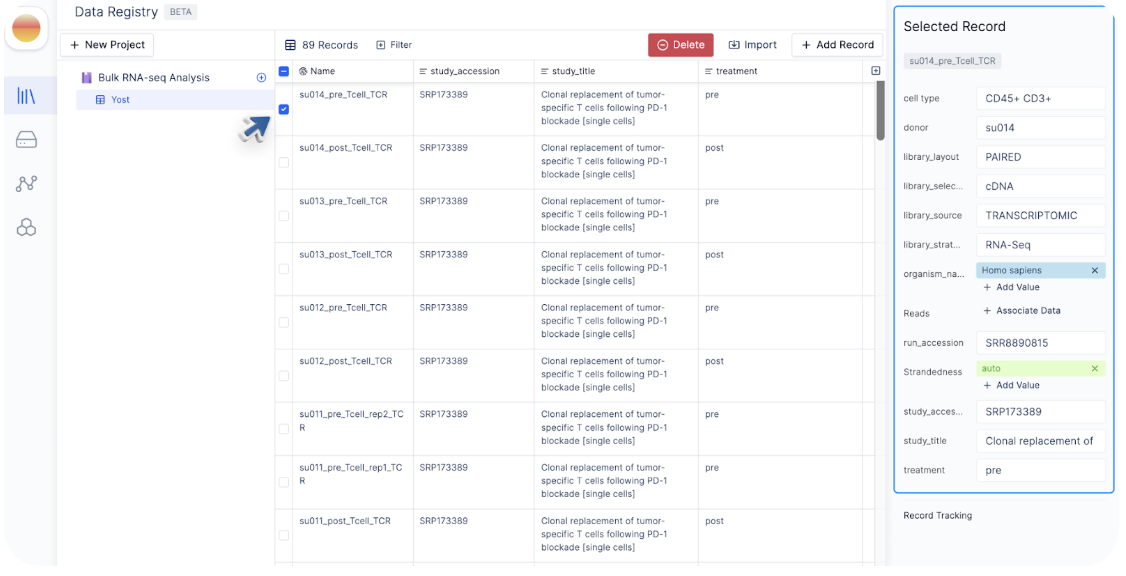
To identify the most suitable antibody candidates, scientists may employ several rounds of phage display assays. To facilitate this, we can create a table consisting of SRA IDs and their corresponding FastQ files in Latch Registry. Each biological sample is accompanied by information regarding the selection method employed for the phage display assay, as well as the experimental conditions. Following data acquisition, this table can be incorporated into a Latch workflow to derive enrichment scores for diverse antibody sequences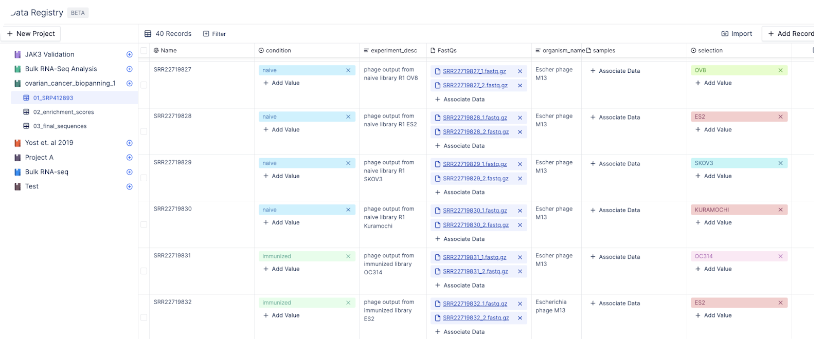
Explore top differentially expressed genes using public transcriptomics studies.
In this project, we imported several transcriptomics studies using bulk RNA-sequencing to identify differentially expressed genes and pathways across a range of biological conditions.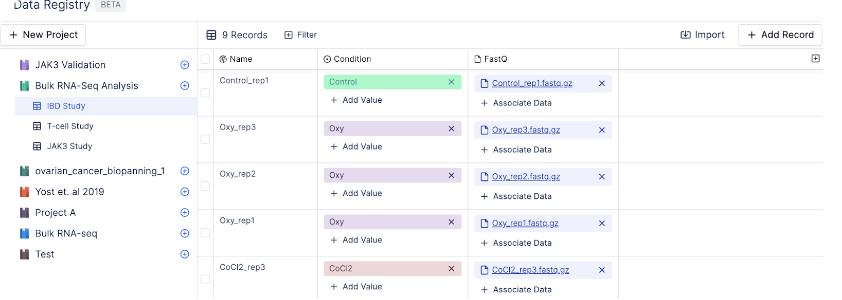
2. Create a Table
What is a table?
What is a table?
A table is a collection of records. Each record, also known as a row, contains a unique record name and a set of attributes.For example, the table IBD Study below contains different sample names, conditions, and sequencing reads.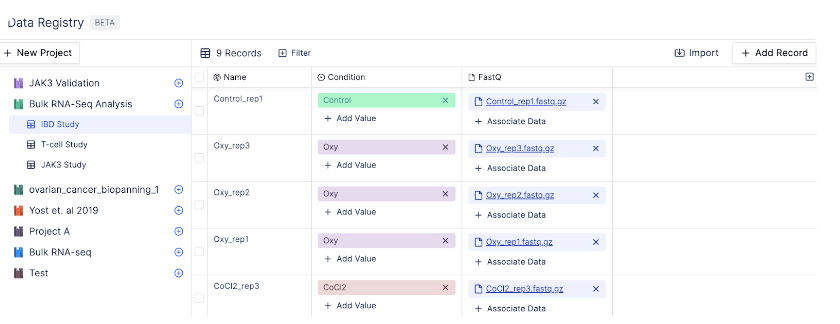

3. Add Records to a Table
You can add records manually, link existing files within latch, import data with a CSV, or sync data from Benchling.Manually Create Single Records
Bulk Import Data Using a CSV
Bulk Link Sequencing Files to Existing Records
Benchling Integration
4. Modify Records in a Table
There are multiple ways you can modify record(s) in a registry table:Edit a single cell value at a time
Similar to a spreadsheet interface, you can double click on each cell and assign it to a new value. Alternatively, you can check the box next to the record you want to modify. The sidebar will display attributes associated with that record and provide an easy way where you can edit.