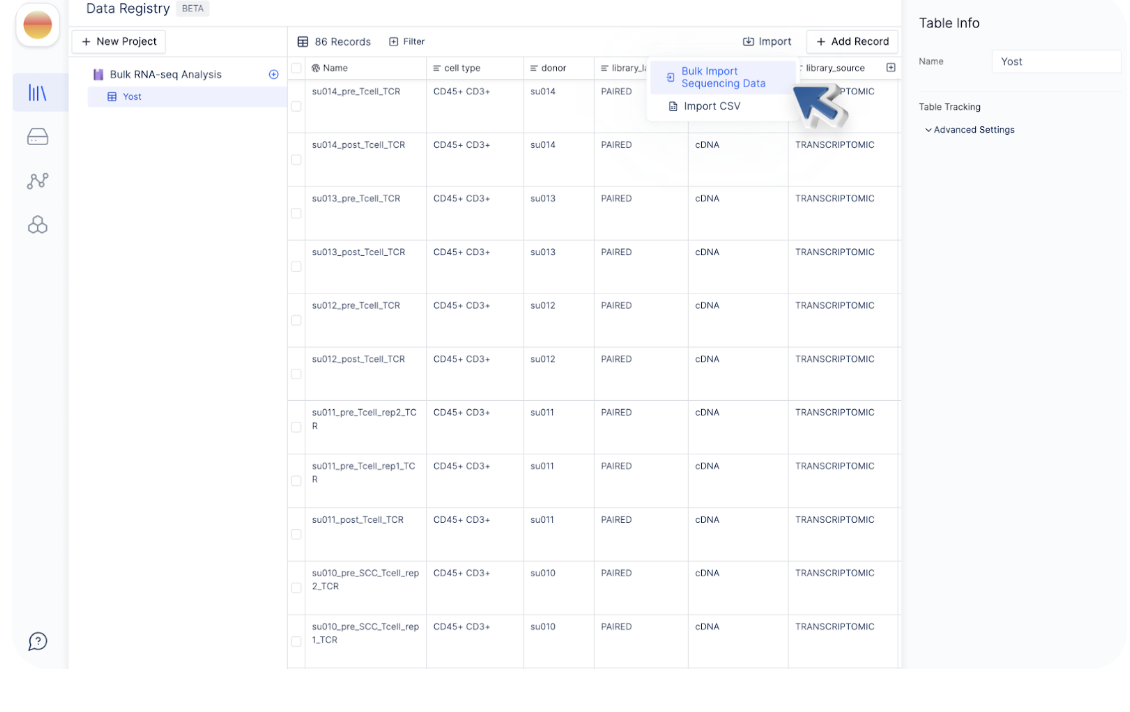
To bulk link sequencing files on Latch with records in registry table, click “Import” and select “Bulk Import Sequencing Data”.
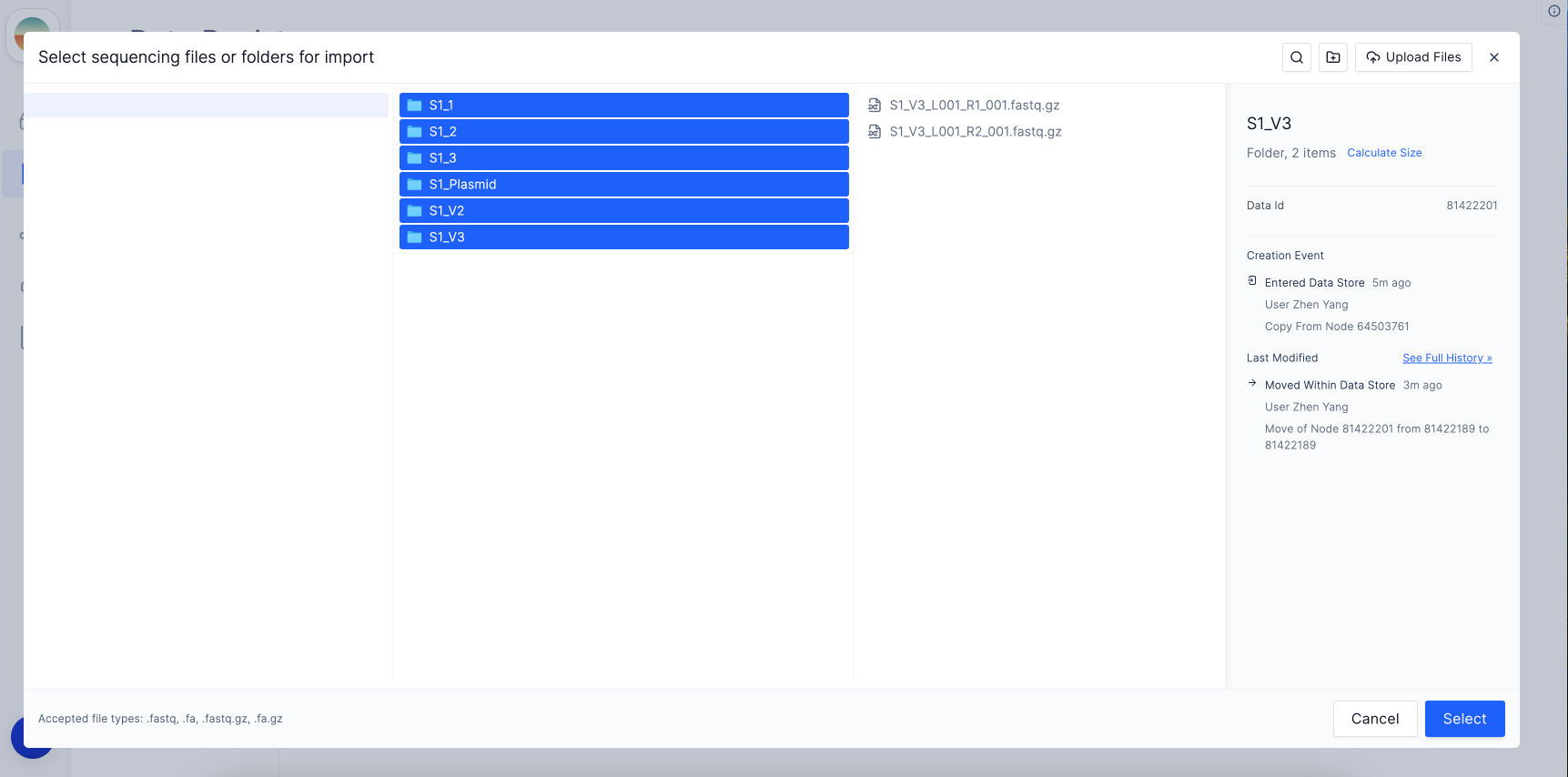
To select multiple folders, click on the first folder, hold Shift on your keyboard, and select the final folder that contains the sequencing files you want to import. Then, choose “Select”.
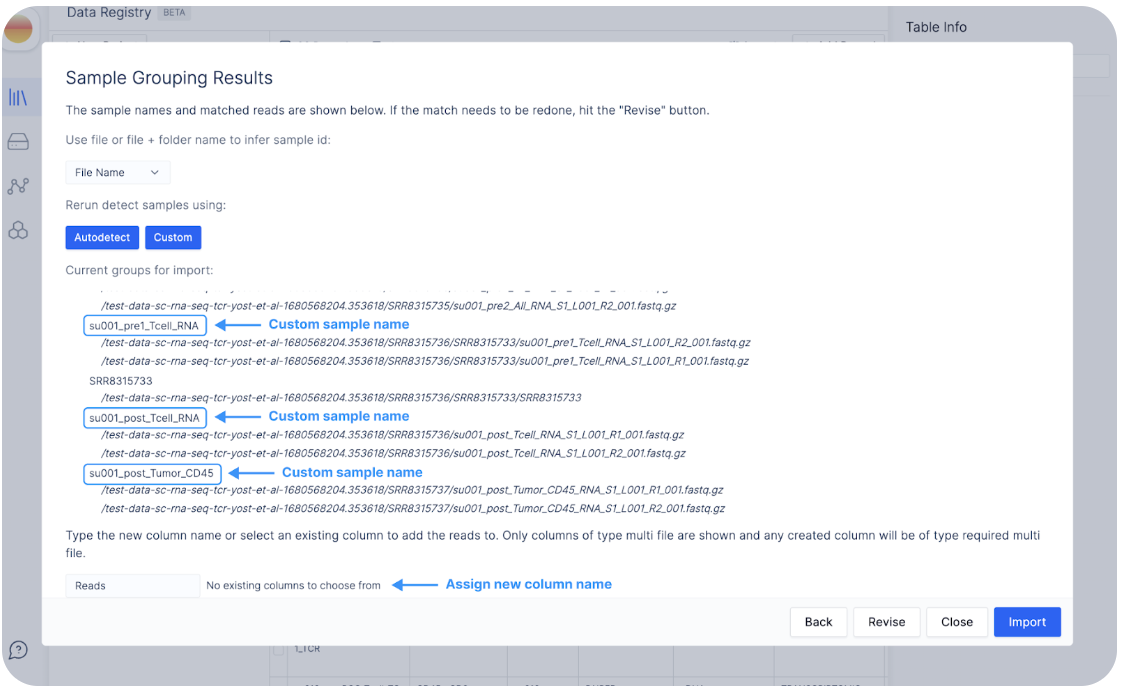
Click on “Autodetect” to see how Latch infers sample names from file names.
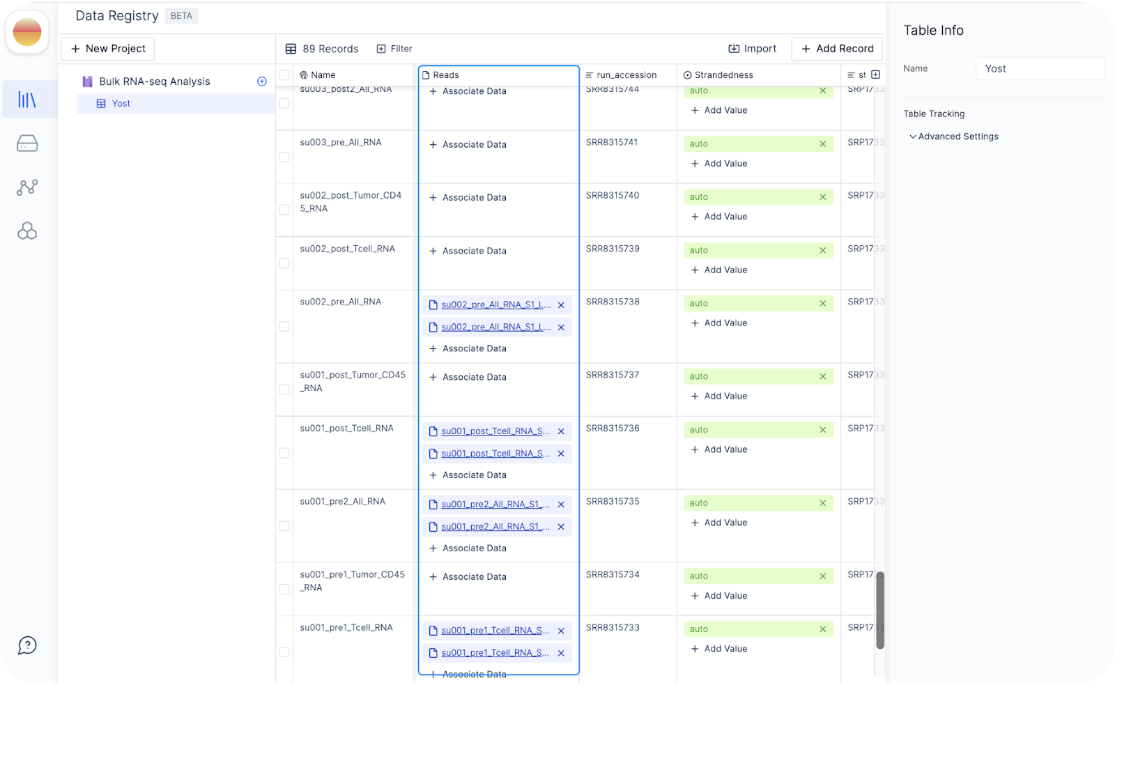
For example, the files 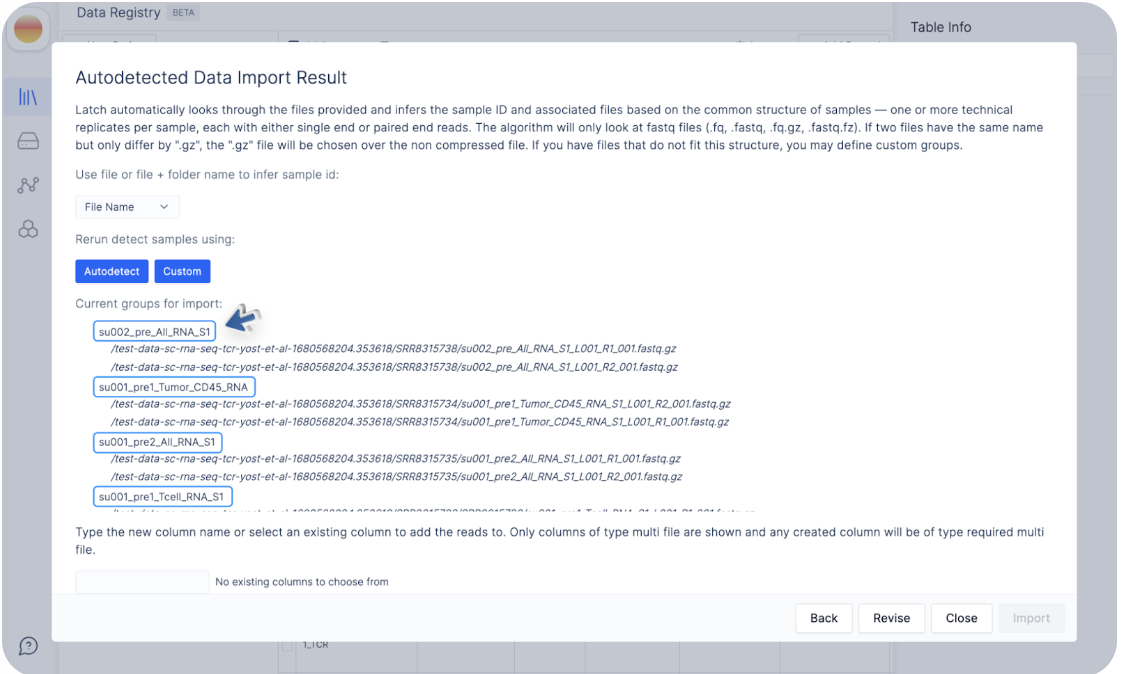
/test-data-sc-rna-seq-tcr-yost-et-al-1680568204.353618/SRR8315738/su002_pre_All_RNA_S1_L001_R1_001.fastq.gz and /test-data-sc-rna-seq-tcr-yost-et-al-1680568204.353618/SRR8315738/su002_pre_All_RNA_S1_L001_R2_001.fastq.gz have the sample names su002_pre_All_RNA_S1 as that is the common prefix between both files.
Let’s say we are not happy with these sample names, and would like the samples to be named `su002_pre_All_RNA` instead of `su002_pre_All_RNA_S1`.
To do so, we can click on “Custom”.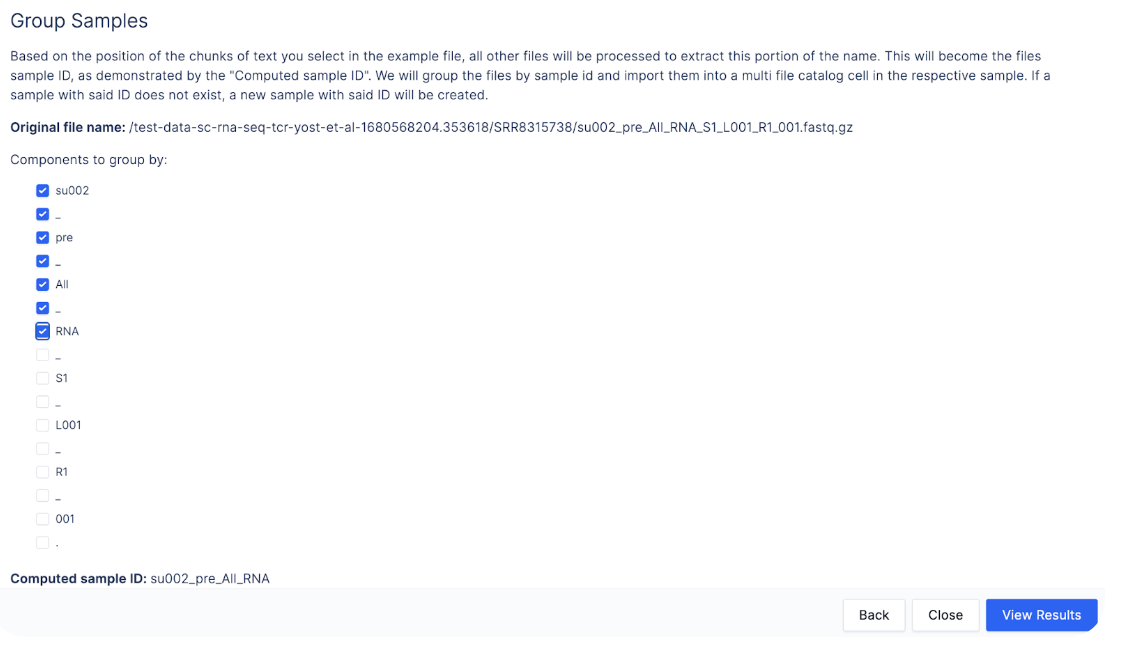

Here, we can check off the boxes from su002 to RNA to create the custom sample name `su002_pre_All_RNA`.
Click “View Results” to see the custom sample names.